Quantum-classical model of excitation transfer along a DNA fragment is computed. This DNA fragment is considered to be a chain of N sites. Each of these sites behaves as a harmonic oscillator. It is conventional, that the base planes of nucleotides are parallel to one another, and the distance between base planes of neighbouring sites are constant; this corresponds to the B-form of DNA.
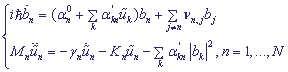 ,
,
where
bn is the probability amplitude of the electron localization at the n -th site,
α0n is the excitation energy at the n -th site,
α'n is coupling constant between the charge with nucleotides,
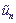 are displacements of sites from their equilibrium positions,
are displacements of sites from their equilibrium positions,
νi,j are matrix elements of transition from the i -th to the j -th site,
Mk is the mass of the k -th site,
γk is the friction coefficient,
Kk is the elastic constant.
All derivatives are differentiated here with time 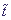 .
.
Let τ = 10-14 sec is the characteristic time,
Un is the characteristic size of fluctuation of the n -th site,
that is 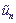 = Unun,
= Unun, 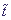 = τ·t.
The corresponding system in proximity of the nearest neighbours in non-dimensional variables has the following form:
= τ·t.
The corresponding system in proximity of the nearest neighbours in non-dimensional variables has the following form:
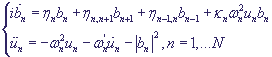 ,
,
The effective mass of all sites is considered to be the same, and it is equal to Mn = 10-21 g.
High frequency intramolecular oscillations in DNA, which correspond to oscillations of bases in a separate site, have frequencies of picosecond order. Frequencies and friction coefficients on sites are considered to be the same; the corresponding non-dimensional values lay in the range
ω2n ≈ 10-4 - 10-6, ω'n ≈ 10-2 - 10-5.
The system is solved by the fourth-order Runge-Kutta method with constant step.