Length
Length is the size of the reliable heterogeneity (latent periodicity) region revealed on a given chromosome of a specified organism. Length is a search field, whereby the user may specify a diapason of lengths for periodicity regions queried in HeteroGenome. On setting a value for parameter Output mode equal to «simple», a search is conducted across all periodicity regions on the given chromosome of the specified organism. On setting Output mode equal to «nonredundant», a search is conducted only across those regions of the first level, i.e., across non-intersected Group representers that form nonredundant coverage of chromosomes by periodicity regions.
Period
The spectral-statistical approach to revealing latent periodicity is based on statistical criteria of testing homogeneity in textual strings (DNA sequences). First, the analyzed chromosome region of length n consisting of characters of the alphabet A = {a1,...,aK} is divided into substrings of length λ, also referred to as the test-period λ. A search for significant heterogeneity in an analyzed textual string (region DNA sequence) is conducted for all possible values of λ for the string test-periods in diapason 2,...n/5K, . Joint use of the spectral-statistical approach parameters – the H-spectrum of heterogeneity manifestation and the
Period is a search field. For a given chromosome of a specified organism, the user may set a diapason of pattern sizes (Periods) for the periodicity regions of interest. For example, if values set in the Period field are greater than 100, mega-satellite regions will be selected. Upon setting a value for the parameter
Group
To present nonredundant data in HeteroGenome, a two-level logical unit record called a Group has been developed. A Group is represented by collocated regions with reliable heterogeneity (latent periodicity) on the chromosome. The longest region in a Group is the Group representer (region of the first level). The remaining regions (regions of the second level) are considered the elements of internal heterogeneity in the representer. Generally, these elements are well-defined structures of periodicity in the DNA sequence of Group representer. Sometimes, additional analysis of Group elements aids a more correct interpretation of data compared to that performed simply by computer. After analysis of Group content, the user may correct the Group borders and even re-evaluate this Group as consisting of a few different Groups.
In addition to Groups containing regions of both levels (Group representer and its internal heterogeneity elements), some Groups consist of only one region of a representer. In general, different Groups do not intersect. Thus, the Group representers form nonredundant coverage of the chromosome by regions of reliable heterogeneity (latent periodicity). Information about each Group in HeteroGenome is shown on a separate web page in the user’s browser. The parameters of Group representer (region of the first level) are presented at the top of page. Below, under the heading INTRINSIC HETEROGENETIES, the parameters of internal heterogeneity elements (regions of the second level) are shown. Data about the region that fit the user’s query are highlighted in red. A scaled graphical scheme of the whole Group reflecting its structural organization is displayed at the bottom of the page.
Output mode
Because of the two-level organization of a logical unit record (Group) in HeteroGenome, data that fit the search parameters may be collected at both levels. To query data only for Group representers (regions of the first level) the value for the parameter Output mode must be equal to that of «nonredundant». On setting the parameter Output mode equal to «simple», data is selected across both levels.
Location
Location is the coordinates of reliable heterogeneity (possible latent periodicity) region on the given chromosome of the specified organism. Location is a search field. Diapason for the coordinates of the periodicity regions on the given chromosome may be specified in this field according to user interest. When Output mode is equal to «simple», a search is performed across all periodicity regions found on the specified chromosome of the given organism. On setting Output mode equal to «nonredundant», a search is conducted across first-level regions only, i.e., across the Group representers, which make up nonredundant chromosome coverage by periodicity regions.
Exponent (RL-value)
RL-value is a real number associated with the number of copies of periodicity pattern in the considered chromosome region of length n. This number indicates how many times a DNA fragment of Period length L may be repeated along the Length n of the chromosome region whose coordinates are specified in the Location field:
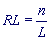 .
.
Exponent is a search field. It enables the user to specify a diapason of a pattern copy number of interest for periodicity regions on the stipulated chromosome of an organism. For example, the user may be interested only in those regions with periodicity pattern copy numbers greater than 10 (RL > 10). When Output mode is equal to «simple», a search is performed across all periodicity regions found on the specified chromosome. On setting Output mode equal to «nonredundant», a search is conducted across first-level regions only, i.e., across the Group representers, which make up nonredundant chromosome coverage by periodicity regions.
Preservation Level (PL-value)
PL-value is the value of the pl-spectrum of a character preservation level in a textual string (an analyzed DNA sequence region) at Period length L. PL-value is the average value for the frequencies of dominant characters in the positions of Period. This parameter estimates average invariance of copies for the periodicity pattern (0.4 ≤ PL ≤ 1). Calculation of PL-value begins with the building up of L-profile.
Preservation Level is a search field. A diapason of PL-value of interest to the user may be specified in HeteroGenome for periodicity regions on the given chromosome of the preset organism. For example, regions with low Preservation Level (0.4 ≤ PL ≤ 0.7) only, which correspond to highly diverged tandem repeats, may be of interest. With Output mode equal to «simple», a search is conducted across all periodicity regions found on the specified chromosome. On setting Output mode equal to «nonredundant», a search is undertaken across first-level regions only, i.e., across the Group representers, which make up nonredundant chromosome coverage by periodicity regions.
L-profile
The analyzed region of textual string (DNA sequence) of length n, consisting of characters from the alphabet A = {a1,...,aK}, is divided into substrings of Period length L, which are placed one on top of another. This array of substrings is called the L-profile, and the number of substrings
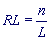 is called the Exponent. L-profile enables calculation of the occurrence frequency
is called the Exponent. L-profile enables calculation of the occurrence frequency 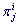 of the ith character from the alphabet A in the jth position (jth column) of this profile. Using frequencies
of the ith character from the alphabet A in the jth position (jth column) of this profile. Using frequencies 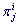 (
(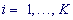 and
and 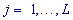 ), the PL-value of the pl-spectrum of character preservation level is calculated at length L.
), the PL-value of the pl-spectrum of character preservation level is calculated at length L.
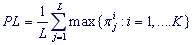 .
.
pl-spectrum
The pl-spectrum of character preservation level over the positions of λ-profile (analog of L-profile with substrings of length λ) for the test-period length λ in a textual string is as follows:
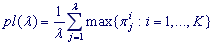 .
.
pl-spectrum amounts to maximum PL-value at length λ = L of the estimated pattern of the latent periodicity region (approximate tandem repeat). For any sequence in HeteroGenome, the pl-spectrum may be viewed on a tab named Show spectra.
HL-value
HL-value is the value of the H-spectrum of heterogeneity
manifestation in a textual string (an analyzed DNA sequence) at Period length L.
To calculate the HL-value corresponding to Period, the L-profile is used,
which enables the frequency 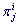 of occurrence of the ith
character from the alphabet A = {a1,...,aK in the jth position (jth column)
of this profile to be determined for
of occurrence of the ith
character from the alphabet A = {a1,...,aK in the jth position (jth column)
of this profile to be determined for 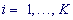 and
and
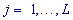 . If pi is a frequency of occurrence of
the ith character from the alphabet A in a whole textual string, then HL-value is calculated at
length L by using normalized Pearson statistics:
. If pi is a frequency of occurrence of
the ith character from the alphabet A in a whole textual string, then HL-value is calculated at
length L by using normalized Pearson statistics:
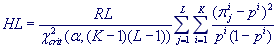 ,
,
where χ2crit is the critical value of χ2-distribution with (K-1)(L-1) degrees of freedom at significance level α = 10-6 and RL is an Exponent for period L.
H-spectrum
H-spectrum of heterogeneity manifestation in a textual string demonstrates clearly the reliable heterogeneity of an analyzed string at the tested λ Period, where H(λ) > 1. The H-spectrum may be viewed on the tab named Show spectra for any sequence in HeteroGenome.
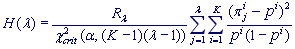 ,
,
where 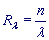 is an Exponent for the tested λ Period.
is an Exponent for the tested λ Period.
Show spectra
For each region of reliable heterogeneity (periodicity) in HeteroGenome, a separate window shows the H-spectrum of heterogeneity manifestation in the region DNA sequence together with the pl-spectrum of character preservation level at tested λ Periods. From joint analysis of these two spectra, the size of the periodicity pattern is estimated. In the upper string of the window from left to right, the following values are listed: region coordinates on a chromosome (Location), pattern size estimate (Period), PL-value corresponding to the pl-spectrum value at the Period length, HL-value corresponding to the H-spectrum value at the same length and RL-value, which is equal to the copy number of the estimated periodicity pattern.
An estimate of periodicity pattern size (Period) is accepted if it meets two conditions. First, the first maximum value in the pl-spectrum is obtained at λ = Period. Second, at λ = Period an inequality H(λ) > 1 is correct for the H-spectrum.
It should be noted that pattern size estimate is deduced automatically from the spectra in the running program. Thus, we suggest that the user analyzes the spectra creatively, paying special attention to the regularity of peaks in both spectra. In some cases an interval length between regular repeating peaks provides the best estimate of periodicity pattern size (Period).
For long spectra, there is an option to view them by parts in the case of a specified diapason of tested λ Periods by pressing the button «Define new range».
Show sequence
For each region of reliable heterogeneity (periodicity), the DNA sequence is shown as a column (L-profile) of consecutive sequence segments with the same length L equal to Period.
The user has the option of changing the size λ for a sequence of segments. On pressing the button «Change» the database generates new sequence segmentation in a separate window. Different visual representations aid correction of Period value for tandem repeats.
Sequence Viewer
For each region of reliable heterogeneity (periodicity), by using the NCBI Sequence Viewer graphical interface (http://www.ncbi.nlm.nih.gov/projects/sviewer/) information on the functional context of an analyzed region on the chromosome can be viewed in a separate window.
References
1. Chaley M., Kutyrkin V. Model of perfect tandem repeat with random pattern and empirical homogeneity testing poly-criteria for latent periodicity revelation in biological sequences. Mathematical Biosciences, 2008, Vol. 211, Issue 1, pp. 186-204.
2. Chaley M.B., Nazipova N.N., Kutyrkin V.A. Statistical Methods for Detecting Latent Periodicity Patterns in Biological Sequences: The Case of Small-Size Samples. Pattern Recognition and Image Analysis. 2009, Vol. 19, No. 2, pp. 358-367.
3. Chaley M., Nazipova N., Teplukhina E., Tyulbasheva G., Kutyrkin V. Statistical Methods for Detecting Latent Periodicity in Biological Sequences: Solving a Problem of Small-Size Samples. In: 2009 IEEE International Conference on Bioinformatics and Biomedicine: the book of abstracts of the BIBM 2009 (Nov 1 – 4, 2009, Washington D.C.). Los Alamitos: IEEE Computer Society, 2009, pp. 92-96.